外泌体共享数据库
Exosome Database
EVpedia-- Database of exosomal proteome, transcriptome, and lipidome
A public database for extracellular vesicles research.
EVpedia is an integrated and comprehensive proteome, transcriptome, and lipidome database of EVs derived from archaea, bacteria, and eukarya, including human.
EVpedia provides an array of tools, such as search and browse tools for vesicular proteins, comparison of vesicular datasets by ortholog identification, Gene Ontology enrichment analyses and network analyses of vesicular proteins. Furthermore, EVpedia provides databases of vesicular mRNAs, miRNAs, and lipids. Thus, EVpedia might serve as a useful community resource to trigger the advancement of systematic and comprehensive studies of EVs and to unveil the fundamental roles of EVs.
ExoCarta--Database of exosomal proteins, RNA and lipids.

ExoCarta: database of exosomal proteins, RNA and lipids.
ExoCarta, an exosome database, provides with the contents that were identified in exosomes in multiple organisms.
Vesiclepedia -- Database of exosomal proteins, RNA and lipids
A community compendium for extracellular vesicles.
Vesiclepedia, a manually curated compendium of molecular data (lipid, RNA and protein) identified in different classes of EVs. Currently, Vesiclepedia comprises 35,264 protein, 18,718 mRNA, 1,772 miRNA and 342 lipid entries encompassed from 341 independent studies that were published over the past several years.
Exosome Gene Ontology Annotation Initiative
http://www.ebi.ac.uk/GOA/exosome
![]()
This initiative, undertaken by the UniProt curators at the EBI, involves the manual assignment of GO annotations to human exosomal proteins by curators reading the scientific literature and assigning to the proteins GO terms that describe their biological roles. Curators working on this project are requesting new GO terms specific to exosome protein biology to fully capture literature findings.
exoRBase --Database of exosomal circRNA、LncRNA and mRNA
exoRBase is a repository of circular RNA (circRNA), long non-coding RNA (lncRNA) and messenger RNA (mRNA) derived from RNA-seq data analyses of human blood exosomes. Experimental validations from published literature are also included.
exoRBase features the integration and visualization of RNA expression profiles based on normalized RNA-seq data spanning both normal individuals and patients with different diseases.
Urinary Exosome Protein Database
https://hpcwebapps.cit.nih.gov/ESBL/Database/Exosome/
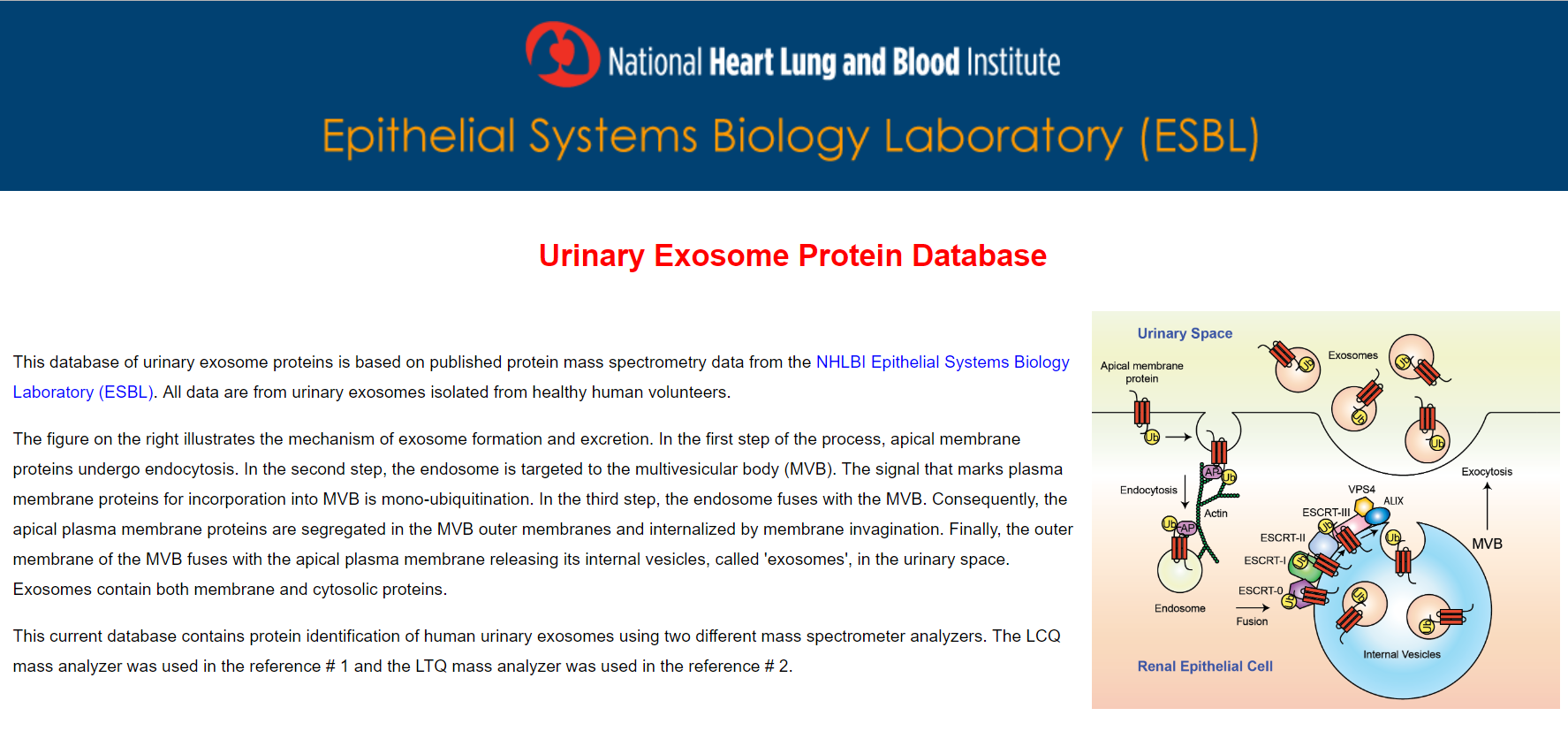
This database of urinary exosome proteins is based on published protein mass spectrometry data from the NHLBI Epithelial Systems Biology Laboratory (ESBL). All data are from urinary exosomes isolated from healthy human volunteers.
This current database contains protein identification of human urinary exosomes using two different mass spectrometer analyzers.
exRNA Atlas--Database of exosomal RNA
The exRNA Atlas is the data repository of the Extracellular RNA Communication Consortium (ERCC), which includes small RNA sequencing and RT-qPCR-derived exRNA profiles from human and mouse biofluids. All RNA-seq datasets are processed using version 4 of the exceRpt small RNA-seq pipeline and ERCC-developed quality metrics are uniformly applied to these datasets.
edited by Alex from Umibio (Shanghai) Co. Ltd